import numpy as np
import pandas as pd1 빅데이터와 금융자료 분석 CH1
missdict = {'f1': [1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
'f2': [10., None, 20., 30., None, 50., 60., 70., 80., 90.],
'f3': ['A', 'A', 'A', 'A', 'B', 'B', 'B', 'B', 'C', 'C']}
missdata = pd.DataFrame( missdict )
missdata.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 10 entries, 0 to 9
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 f1 10 non-null int64
1 f2 8 non-null float64
2 f3 10 non-null object
dtypes: float64(1), int64(1), object(1)
memory usage: 368.0+ bytesmissdata.isna().mean()f1 0.0
f2 0.2
f3 0.0
dtype: float64tmpdata1 = missdata.dropna()
tmpdata1| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 1 | 10.0 | A |
| 2 | 3 | 20.0 | A |
| 3 | 4 | 30.0 | A |
| 5 | 6 | 50.0 | B |
| 6 | 7 | 60.0 | B |
| 7 | 8 | 70.0 | B |
| 8 | 9 | 80.0 | C |
| 9 | 10 | 90.0 | C |
tmpdata2 = missdata.dropna( subset=['f3'] )
tmpdata2| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 1 | 10.0 | A |
| 1 | 2 | NaN | A |
| 2 | 3 | 20.0 | A |
| 3 | 4 | 30.0 | A |
| 4 | 5 | NaN | B |
| 5 | 6 | 50.0 | B |
| 6 | 7 | 60.0 | B |
| 7 | 8 | 70.0 | B |
| 8 | 9 | 80.0 | C |
| 9 | 10 | 90.0 | C |
numdata = missdata.select_dtypes(include=['int64', 'float64'])
tmpdata3 = numdata.fillna( -999, inplace=False )
tmpdata3.describe()| f1 | f2 | |
|---|---|---|
| count | 10.00000 | 10.000000 |
| mean | 5.50000 | -158.800000 |
| std | 3.02765 | 443.562297 |
| min | 1.00000 | -999.000000 |
| 25% | 3.25000 | 12.500000 |
| 50% | 5.50000 | 40.000000 |
| 75% | 7.75000 | 67.500000 |
| max | 10.00000 | 90.000000 |
numdata.mean()f1 5.50
f2 51.25
dtype: float64tmpdata4 = numdata.fillna( numdata.mean(), inplace=False )
tmpdata4| f1 | f2 | |
|---|---|---|
| 0 | 1 | 10.00 |
| 1 | 2 | 51.25 |
| 2 | 3 | 20.00 |
| 3 | 4 | 30.00 |
| 4 | 5 | 51.25 |
| 5 | 6 | 50.00 |
| 6 | 7 | 60.00 |
| 7 | 8 | 70.00 |
| 8 | 9 | 80.00 |
| 9 | 10 | 90.00 |
missdata.groupby('f3')['f2'].mean()f3
A 20.0
B 60.0
C 85.0
Name: f2, dtype: float64missdata.groupby('f3')['f2'].transform('mean')0 20.0
1 20.0
2 20.0
3 20.0
4 60.0
5 60.0
6 60.0
7 60.0
8 85.0
9 85.0
Name: f2, dtype: float64tmpdata5 = numdata.copy()
tmpdata5['f2'].fillna( missdata.groupby('f3')['f2'].transform('mean'), inplace=True)
tmpdata5/var/folders/n2/jbh_0_091bx8qgz7j87t2qwc0000gp/T/ipykernel_25894/622840210.py:2: FutureWarning: A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
tmpdata5['f2'].fillna( missdata.groupby('f3')['f2'].transform('mean'),inplace=True)| f1 | f2 | |
|---|---|---|
| 0 | 1 | 10.0 |
| 1 | 2 | 20.0 |
| 2 | 3 | 20.0 |
| 3 | 4 | 30.0 |
| 4 | 5 | 60.0 |
| 5 | 6 | 50.0 |
| 6 | 7 | 60.0 |
| 7 | 8 | 70.0 |
| 8 | 9 | 80.0 |
| 9 | 10 | 90.0 |
missdata_tr = missdata.dropna()
x_tr = missdata_tr[['f1']]
y_tr = missdata_tr['f2']
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit( x_tr, y_tr )
missdata_ts = missdata [ missdata.isnull().any(axis=1) ]
x_ts = missdata_ts[['f1']]
predicted_values = model.predict( x_ts )
tmpdata6 = missdata.copy()
tmpdata6.loc[ tmpdata6['f2'].isnull(), 'f2'] = predicted_values
tmpdata6| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 1 | 10.000000 | A |
| 1 | 2 | 14.191176 | A |
| 2 | 3 | 20.000000 | A |
| 3 | 4 | 30.000000 | A |
| 4 | 5 | 41.985294 | B |
| 5 | 6 | 50.000000 | B |
| 6 | 7 | 60.000000 | B |
| 7 | 8 | 70.000000 | B |
| 8 | 9 | 80.000000 | C |
| 9 | 10 | 90.000000 | C |
missdata_num = missdata.copy()
missdata_num['f3']=missdata_num['f3'].map({'A':1,'B':2,'C':3})missdata_num| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 1 | 10.0 | 1 |
| 1 | 2 | NaN | 1 |
| 2 | 3 | 20.0 | 1 |
| 3 | 4 | 30.0 | 1 |
| 4 | 5 | NaN | 2 |
| 5 | 6 | 50.0 | 2 |
| 6 | 7 | 60.0 | 2 |
| 7 | 8 | 70.0 | 2 |
| 8 | 9 | 80.0 | 3 |
| 9 | 10 | 90.0 | 3 |
from sklearn.impute import KNNImputer
imputer = KNNImputer(n_neighbors=2)
tmpdata7 = imputer.fit_transform(missdata_num)pd.DataFrame( tmpdata7 )| 0 | 1 | 2 | |
|---|---|---|---|
| 0 | 1.0 | 10.0 | 1.0 |
| 1 | 2.0 | 15.0 | 1.0 |
| 2 | 3.0 | 20.0 | 1.0 |
| 3 | 4.0 | 30.0 | 1.0 |
| 4 | 5.0 | 40.0 | 2.0 |
| 5 | 6.0 | 50.0 | 2.0 |
| 6 | 7.0 | 60.0 | 2.0 |
| 7 | 8.0 | 70.0 | 2.0 |
| 8 | 9.0 | 80.0 | 3.0 |
| 9 | 10.0 | 90.0 | 3.0 |
outdict = {'A': [10, 0.02, 0.3, 40, 50, 60, 712, 80, 90, 1003],
'B': [0.05, 0.00015, 25, 35, 45, 205, 65, 75, 85, 3905]}
outdata = pd.DataFrame( outdict )
Q1 = outdata.quantile(0.25)
Q3 = outdata.quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
((outdata < lower_bound) | (outdata > upper_bound))| A | B | |
|---|---|---|
| 0 | False | False |
| 1 | False | False |
| 2 | False | False |
| 3 | False | False |
| 4 | False | False |
| 5 | False | True |
| 6 | True | False |
| 7 | False | False |
| 8 | False | False |
| 9 | True | True |
outliers = ((outdata < lower_bound) | (outdata > upper_bound)).any(axis=1)
outliersdata = outdata[ outliers ]
outliersdata| A | B | |
|---|---|---|
| 5 | 60.0 | 205.0 |
| 6 | 712.0 | 65.0 |
| 9 | 1003.0 | 3905.0 |
standardizeddata = (outdata - outdata.mean()) / outdata.std()
standardizeddata| A | B | |
|---|---|---|
| 0 | -0.552206 | -0.364647 |
| 1 | -0.580536 | -0.364688 |
| 2 | -0.579741 | -0.344154 |
| 3 | -0.467047 | -0.335940 |
| 4 | -0.438661 | -0.327727 |
| 5 | -0.410274 | -0.196309 |
| 6 | 1.440519 | -0.311300 |
| 7 | -0.353501 | -0.303086 |
| 8 | -0.325115 | -0.294872 |
| 9 | 2.266563 | 2.842723 |
outliers2 = ((standardizeddata < -3) | (standardizeddata > 3)).any(axis=1)
outliersdata2 = outdata[ outliers2 ]
outliersdata2| A | B |
|---|
import matplotlib.pyplot as plt
np.random.seed(42)
X_inliers = 0.3 * np.random.randn(100, 2)
X_outliers = np.random.uniform(low=-4, high=4, size=(20, 2))
X = np.r_[X_inliers + 2, X_inliers - 2, X_outliers]
plt.figure(figsize=(5, 4))
plt.scatter(X[:, 0], X[:, 1], color='k', s=20)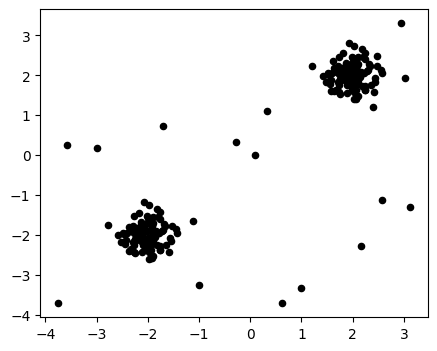
from sklearn.neighbors import LocalOutlierFactor
clf = LocalOutlierFactor(n_neighbors=20, contamination=0.1)
y_pred = clf.fit_predict(X) # 1: inlier, -1: outlier
outlier_mask = y_pred == -1
plt.figure(figsize=(5, 4))
plt.scatter(X[:, 0], X[:, 1], color='b', s=20, label='Inliers')
plt.scatter(X[outlier_mask, 0], X[outlier_mask, 1], color='r', s=50,label='Outliers')
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.legend()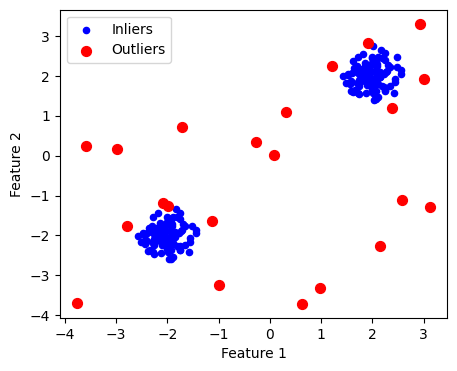
from sklearn.ensemble import IsolationForest
clf2 = IsolationForest(contamination=0.1)
# contamination : 이상치 비율
# n_estimators : 나무의 갯수 (defalut 100)
# max_features : 각 나무별 특성변수의 갯수(default 1)
clf2.fit( X )
y_pred2 = clf2.predict( X ) # 1: inlier, -1: outlier
outlier_mask2 = y_pred2 == -1
plt.figure(figsize=(5, 4))
plt.scatter(X[:, 0], X[:, 1], color='b', s=20, label='Inliers')
plt.scatter(X[outlier_mask2, 0], X[outlier_mask2, 1], color='r', s=50,label='Outliers')
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.legend()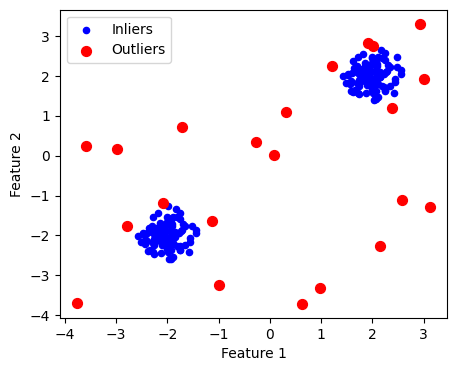
clf2.score_samples(X)array([-0.39567479, -0.43973362, -0.38576701, -0.4625295 , -0.39169087,
-0.39353101, -0.44632054, -0.4539587 , -0.39410011, -0.42823486,
-0.43802322, -0.4231585 , -0.38600184, -0.40324911, -0.38778406,
-0.46849881, -0.4075734 , -0.43420431, -0.45140279, -0.4113938 ,
-0.40036102, -0.38335266, -0.42666089, -0.41104579, -0.44476313,
-0.38744281, -0.39942914, -0.44257912, -0.38938554, -0.40470075,
-0.39006555, -0.42881353, -0.44275864, -0.40154039, -0.39729665,
-0.42434279, -0.42743206, -0.57699503, -0.38628797, -0.45454533,
-0.3864026 , -0.44513991, -0.39598029, -0.41231495, -0.39270763,
-0.40568073, -0.39005843, -0.43210962, -0.38601781, -0.38485125,
-0.41991455, -0.40181793, -0.38788591, -0.48400217, -0.38538727,
-0.47693547, -0.50815903, -0.39115267, -0.40423505, -0.43216634,
-0.4254748 , -0.48475901, -0.4890737 , -0.40357175, -0.39439224,
-0.42406868, -0.40171635, -0.44870204, -0.38761922, -0.43170548,
-0.42364173, -0.43593615, -0.39560581, -0.4391784 , -0.39543452,
-0.38550106, -0.38910014, -0.39558501, -0.48693408, -0.41865632,
-0.40964165, -0.43730378, -0.41716448, -0.47893783, -0.39791987,
-0.40492088, -0.38431516, -0.39544321, -0.42208103, -0.53522817,
-0.41839783, -0.40171635, -0.39362168, -0.39333773, -0.44128807,
-0.40208128, -0.40907841, -0.39096216, -0.38929625, -0.40645206,
-0.38953796, -0.43147334, -0.38691831, -0.45292849, -0.39365031,
-0.40064735, -0.46513506, -0.48209563, -0.39635657, -0.44569517,
-0.4496465 , -0.43243444, -0.39406682, -0.40625773, -0.39826724,
-0.46462545, -0.40782229, -0.42572527, -0.46599338, -0.42852596,
-0.40082304, -0.38971614, -0.4628486 , -0.4219109 , -0.46365237,
-0.39237271, -0.40320941, -0.42432754, -0.40309339, -0.40204058,
-0.39044555, -0.43501511, -0.4324525 , -0.40503508, -0.40259674,
-0.42956023, -0.42799201, -0.56257644, -0.38875652, -0.47585014,
-0.3835507 , -0.4556408 , -0.406217 , -0.40385118, -0.39458774,
-0.40122018, -0.40401747, -0.4443972 , -0.38320086, -0.3834212 ,
-0.44945534, -0.4060406 , -0.38476589, -0.46817324, -0.38466101,
-0.47610349, -0.49275615, -0.38344594, -0.40974845, -0.42395885,
-0.42189161, -0.49261883, -0.48518689, -0.40901991, -0.39452123,
-0.44785724, -0.40375287, -0.45749968, -0.40496115, -0.4260838 ,
-0.41810669, -0.44560803, -0.38806155, -0.45523273, -0.38817405,
-0.38177641, -0.39724261, -0.40279204, -0.46677259, -0.42162856,
-0.4086809 , -0.43961327, -0.40404401, -0.45862325, -0.40473907,
-0.41509113, -0.38081908, -0.39036169, -0.42441162, -0.52130968,
-0.41557892, -0.40347146, -0.39318444, -0.38921027, -0.46342999,
-0.39756969, -0.41357841, -0.38429305, -0.40297782, -0.40881293,
-0.60259641, -0.44411547, -0.57052254, -0.50247903, -0.73212838,
-0.64786135, -0.55623141, -0.45375541, -0.68754218, -0.67833274,
-0.70698798, -0.63422967, -0.64031155, -0.78183101, -0.65477657,
-0.67178443, -0.67084008, -0.68580391, -0.71446705, -0.64861428])